VoxTell: Free-Text Promptable Universal 3D Medical Image Segmentation

Model Description
VoxTell is a state-of-the-art 3D vision-language segmentation model that directly maps free-form text prompts to volumetric masks in medical imaging. Unlike traditional segmentation models that require predefined class labels, VoxTell accepts natural language descriptions ranging from single words to full clinical sentences, enabling intuitive and flexible medical image analysis.
The model is designed for both anatomical and pathological structures across multiple imaging modalities (CT, PET, MRI), being trained on 1000+ familiar concepts while maintaining strong generalization to related unseen classes through its multi-stage vision-language fusion architecture.
Key Features
- Free-text prompting: Generate 3D segmentation masks using natural language descriptions
- Multi-modality support: Works across CT, PET, and MRI imaging modalities
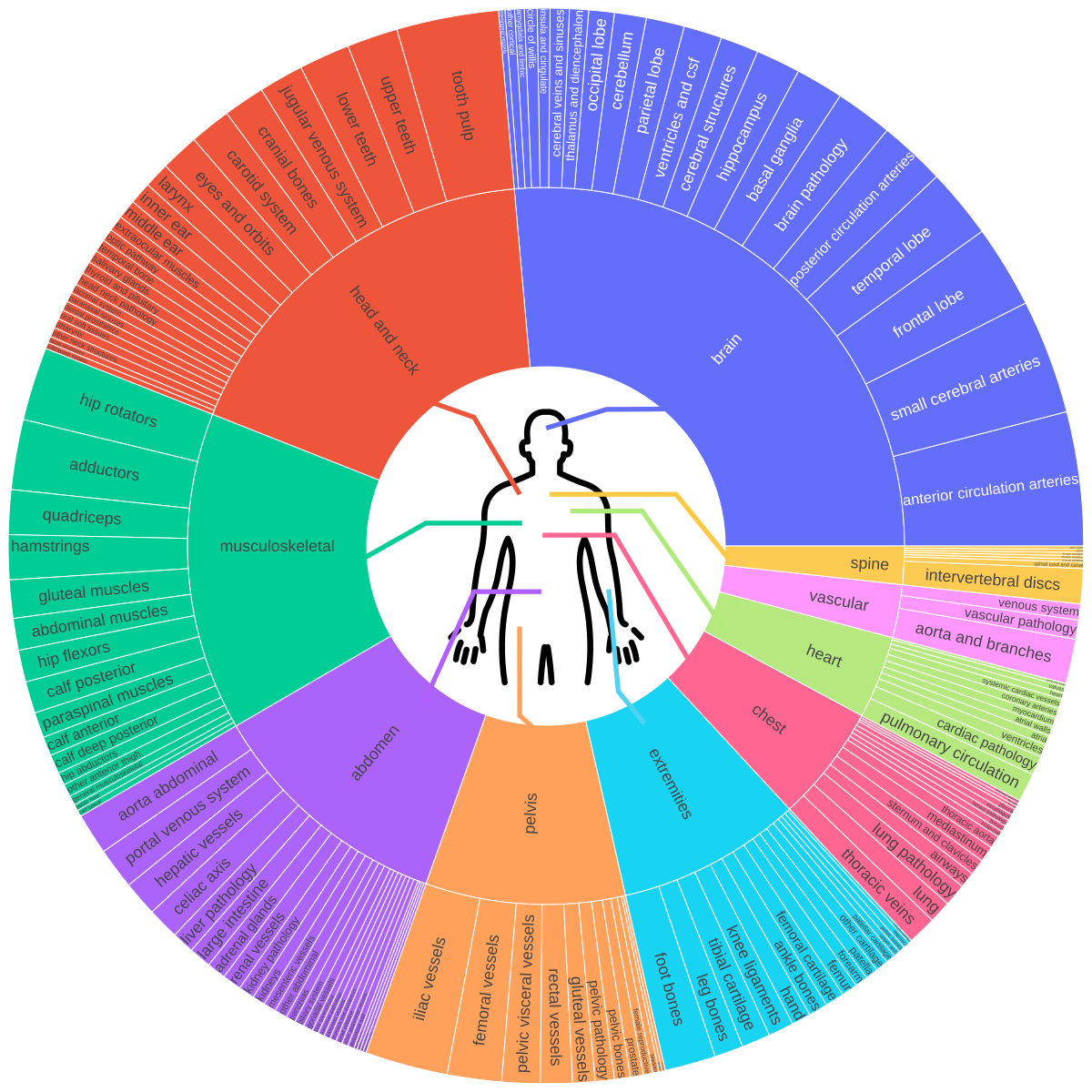
- Comprehensive anatomy coverage: Brain, thorax, abdomen, pelvis, musculoskeletal system, and extremities
- Flexible granularity: From coarse anatomical labels to fine-grained pathological findings
Versions
We release multiple VoxTell versions (continuously updated) to enable both reproducible research and high-performance downstream applications.
VoxTell v1.1 (Recommended)
- Info: This is the current default version
- Training Data: Trained on all datasets from the paper and additional sources (190 datasets, ~68,500 volumes)
- Split: Includes the test sets from the paper in the training corpus
- Sampling Strategy:
- 95% probability: Semantic datasets corpus
- 5% probability: Image-text-mask triplets from instance-focused datasets
- Use Case: Recommended for general application, inference, and fine-tuning. This version maximizes supervision and concept coverage for stronger general-purpose performance
VoxTell v1.0 (Deprecated)
- Info: This version was used for the experiments in the paper but contains known issues that have been fixed in v1.1. It is not recommended for general use.
- Training Data: Trained on 158 datasets (~62,000 volumes)
- Split: Maintains strict train/test separation as described in the paper
- Use Case: Reproducibility of the results reported in the paper
How to Download
You can download VoxTell checkpoints using the Hugging Face huggingface_hub library:
from huggingface_hub import snapshot_download
MODEL_NAME = "voxtell_v1.1" # Updated models may be available in the future
DOWNLOAD_DIR = "/home/user/temp" # Optionally specify the download directory
download_path = snapshot_download(
repo_id="mrokuss/VoxTell",
allow_patterns=[f"{MODEL_NAME}/*", "*.json"],
local_dir=DOWNLOAD_DIR
)
🛠 Installation
1. Create a Virtual Environment
VoxTell supports Python 3.10+ and works with Conda, pip, or any other virtual environment manager. Here's an example using Conda:
conda create -n voxtell python=3.12
conda activate voxtell
2. Install PyTorch
Temporary Compatibility Warning
There is a known issue with PyTorch 2.9.0 causing OOM errors during inference inVoxTell(related to 3D convolutions — see the PyTorch issue here).
Until this is resolved, please use PyTorch 2.8.0 or earlier.
Install PyTorch compatible with your CUDA version. For example, for Ubuntu with a modern NVIDIA GPU:
pip install torch==2.8.0 torchvision==0.23.0 --index-url https://download.pytorch.org/whl/cu126
For other configurations (macOS, CPU, different CUDA versions), please refer to the PyTorch Get Started page.
Install via pip (you can also use uv):
pip install voxtell
or install directly from the GitHub repository:
git clone https://github.com/MIC-DKFZ/VoxTell
cd VoxTell
pip install -e .
3. Python API
For more control or integration into Python workflows, use the Python API:
import torch
from voxtell.inference.predictor import VoxTellPredictor
from nnunetv2.imageio.nibabel_reader_writer import NibabelIOWithReorient
# Select device
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# Load image
image_path = "/path/to/your/image.nii.gz"
img, _ = NibabelIOWithReorient().read_images([image_path])
# Define text prompts
text_prompts = ["liver", "right kidney", "left kidney", "spleen"]
# Initialize predictor
predictor = VoxTellPredictor(
model_dir="/path/to/voxtell_model_directory",
device=device,
)
# Run prediction
# Output shape: (num_prompts, x, y, z)
voxtell_seg = predictor.predict_single_image(img, text_prompts)
4. Optional: Visualize Results
You can visualize the segmentation results using napari:
pip install napari[all]
import napari
import numpy as np
# Create a napari viewer and add the original image
viewer = napari.Viewer()
viewer.add_image(img, name='Image')
# Add segmentation results as label layers for each prompt
for i, prompt in enumerate(text_prompts):
viewer.add_labels(voxtell_seg[i].astype(np.uint8), name=prompt)
# Run napari
napari.run()
Important: Image Orientation and Spacing
⚠️ Image Orientation (Critical): For correct anatomical localization (e.g., distinguishing left from right), images must be in RAS orientation. VoxTell was trained on data reoriented using this specific reader. Orientation mismatches can be a source of error. An easy way to test for this is if a simple prompt like "liver" fails and segments parts of the spleen instead. Make sure your image metadata is correct.
Image Spacing: The model does not resample images to a standardized spacing for faster inference. Performance may degrade on images with very uncommon voxel spacings (e.g., super high-resolution brain MRI). In such cases, consider resampling the image to a more typical clinical spacing (e.g., 1.5×1.5×1.5 mm³) before segmentation.
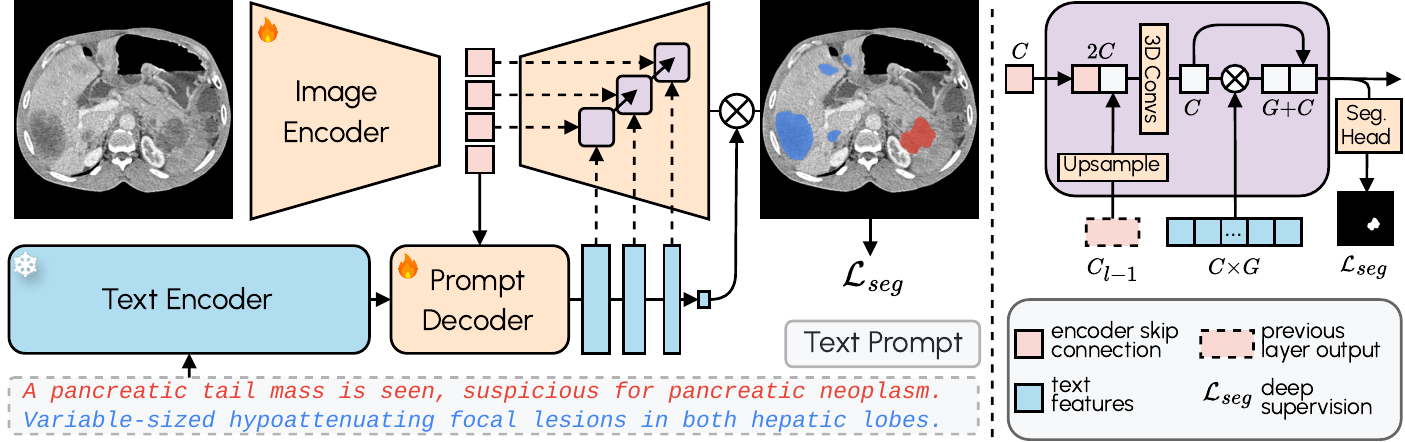
Architecture
VoxTell employs a multi-stage vision-language fusion approach:
- Image Encoder: Processes 3D volumetric input into latent feature representations
- Prompt Encoder: We use the fozen Qwen3-Embedding-4B model to embed text prompts
- Prompt Decoder: Transforms text queries and image latents into multi-scale text features
- Image Decoder: Fuses visual and textual information at multiple resolutions using MaskFormer-style query-image fusion with deep supervision
Intended Use
Primary Use Cases
- Research in vision-language models for medical image analysis
- Text-promptable or automated segmentation of anatomical structures in medical imaging
- Identification and delineation of pathological findings
Out-of-Scope Use
- Clinical diagnosis without expert radiologist review
- Real-time emergency medical decision-making
- Commercial use
Performance
VoxTell achieves state-of-the-art performance on anatomical and pathological segmentation tasks across multiple medical imaging benchmarks. Detailed performance metrics and comparisons are available in the paper.
Tip: Experiment with different prompts tailored to your use case. For example, the prompt lesions is known to be overconfident, i.e. over-segmenting, compared to lesion.
Limitations / Known Issues
- Performance may vary on imaging modalities or anatomical regions underrepresented in training data
- Prompting structures absent from the image and never seen on this modality (e.g., "liver" in a brain MRI) may lead to undesired results
- Text prompt quality and specificity affects segmentation accuracy
- Not validated for direct clinical use without expert review
Citation
@misc{rokuss2025voxtell,
title={VoxTell: Free-Text Promptable Universal 3D Medical Image Segmentation},
author={Maximilian Rokuss and Moritz Langenberg and Yannick Kirchhoff and Fabian Isensee and Benjamin Hamm and Constantin Ulrich and Sebastian Regnery and Lukas Bauer and Efthimios Katsigiannopulos and Tobias Norajitra and Klaus Maier-Hein},
year={2025},
eprint={2511.11450},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2511.11450},
}
📬 Contact
For questions, issues, or collaborations, please contact:
📧 maximilian.rokuss@dkfz-heidelberg.de / moritz.langenberg@dkfz-heidelberg.de
- Downloads last month
- 33



